2D material: h-BN sheet: Difference between revisions
(Created page with "== Prerequisites == thumb|Atomic structure of 2D hBN '''Material properties''': * HCP lattice, ABAB stacking * Four atoms per cell, B and N (16 electrons, )...") |
No edit summary |
||
| Line 1: | Line 1: | ||
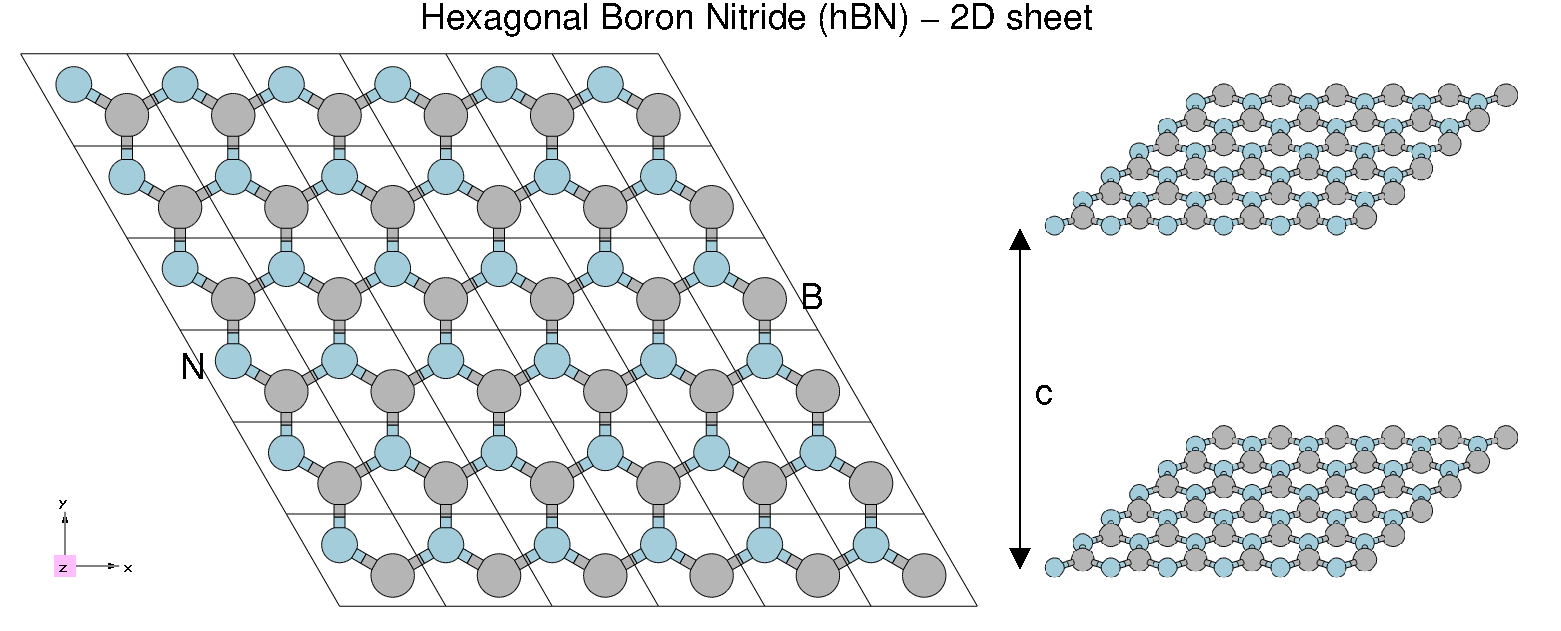
[[File:HBN2.png|thumb|Atomic structure of 2D hBN]] | [[File:HBN2.png|thumb|Atomic structure of 2D hBN]] | ||
In this tutorial you will learn how to generate the Yambo SAVE folder for bulk hBN starting from a PWscf calculation. | |||
== Prerequisites == | |||
'''You will need''': | '''You will need''': | ||
* PWSCF input files and pseudopotentials for hBN bulk (Download here) | * PWSCF input files and pseudopotentials for hBN bulk (Download here) | ||
| Line 13: | Line 9: | ||
* <code>p2y</code> executable | * <code>p2y</code> executable | ||
Unpack the | == System characteristics == | ||
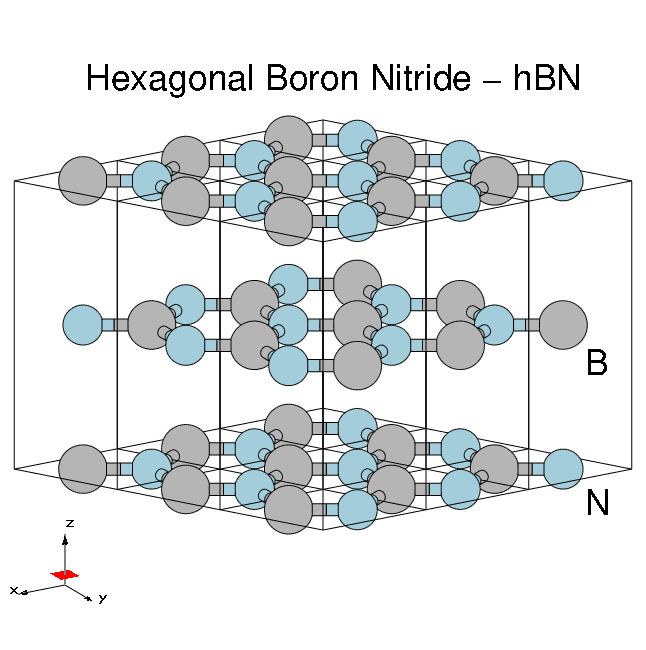
[[File:HBN-bulk-3x3-annotated.png|thumb|Atomic structure of bulk hBN]] | |||
'''Hexagonal boron nitride - hBN''': | |||
* HCP lattice, ABAB stacking | |||
* Four atoms per cell, B and N (16 electrons) | |||
* Lattice constants: ''a'' = 4.716 [a.u.], ''c/a'' = 2.582 | |||
* Plane wave cutoff 40 Ry (1500 RL vectors in wavefunctions) | |||
* SCF run: shifted ''6x6x6'' grid with 8 bands | |||
* Non-SCF run: unshifted ''6x6x2'' grid with 100 bands | |||
== DFT calculations == | |||
Unpack the tarfile. It uses the same file structure as other yambo tutorials: | |||
$ tar -xcvf hBN-bulk.tar | $ tar -xcvf hBN-bulk.tar | ||
$ cd hBN/PWSCF | $ cd YAMBO_TUTORIALS/hBN/PWSCF | ||
$ ls | $ ls | ||
Inputs Pseudos PostProcessing References | |||
hBN_scf.in hBN_nscf.in hBN_scf_plot_bands.in hBN_nscf_plot_bands.in | |||
First run the SCF calculation to generate the ground-state charge density, occupations, Fermi level, and so on: | |||
First run the SCF calculation | |||
pw.x < hBN_scf.in > hBN_scf.out | pw.x < hBN_scf.in > hBN_scf.out | ||
The output reports 36 k-points. The valence band maximum is at 5.13eV. | |||
Next run a non-SCF calculation to generate a set of Kohn-Sham eigenvalues and eigenvectors across a denser k-point mesh, for occupied and unoccupied states: | |||
pw.x < hBN_nscf.in > hBN_nscf.out | pw.x < hBN_nscf.in > hBN_nscf.out | ||
Note the presence of the following flags in the input file: | Note the presence of the following flags in the input file: | ||
wf_collect=.true. | wf_collect=.true. | ||
force_symmorphic=.true. | force_symmorphic=.true. | ||
which are needed for the | which are needed for generating the Yambo databases. Full explanations of these variables are given on the [http://www.quantum-espresso.org/wp-content/uploads/Doc/INPUT_PW.html quantum-ESPRESSO input variables page]. | ||
After these two runs, you should have a <code>hBN.save</code> directory: | |||
$ ls hBN.save | $ ls hBN.save | ||
data-file.xml | data-file.xml charge-density.dat gvectors.dat B.pz-vbc.UPF N.pz-vbc.UPF | ||
K00001 K00002 .... K00035 K00036 | |||
== Conversion to Yambo format == | == Conversion to Yambo format == | ||
PWscf output is converted to the Yambo format using the <code>p2y</code> (pwscf to yambo), found in the yambo <code>bin</code> directory. | The PWscf <code>bBN.save</code> output is converted to the Yambo format using the <code>p2y</code> executable (pwscf to yambo), found in the yambo <code>bin</code> directory. | ||
Enter | Enter <code>hBN.save</code> and launch <code>p2y</code>: | ||
$ cd hBN.save | $ cd hBN.save | ||
$ p2y | $ p2y | ||
... | |||
<---> DBs path set to . | |||
<---> Index file set to data-file.xml | |||
<---> Header/K-points/Energies... done | |||
... | |||
<---> == DB1 (Gvecs and more) ... | |||
<---> ... Database done | |||
<---> == DB2 (wavefunctions) ... done == | |||
<---> == DB3 (PseudoPotential) ... done == | |||
<---> == P2Y completed == | |||
This output repeats some information about the system and generates a <code>SAVE</code> directory: | |||
$ ls SAVE | $ ls SAVE | ||
ns.db1 ns.wf ns.kb_pp_pwscf | ns.db1 ns.wf ns.kb_pp_pwscf | ||
ns.wf_fragments_1_1 ... | ns.wf_fragments_1_1 ... | ||
ns.kb_pp_pwscf_fragment_1 ... | ns.kb_pp_pwscf_fragment_1 ... | ||
These files, with an ''n'' prefix, indicate that they are in netCDF format, and thus not human readable. However, they are perfectly transferable across different architectures. | |||
You are now ready to run Yambo. Check that the databases contain the information you expect: | |||
$ yambo -D | |||
[RD./SAVE//ns.db1]------------------------------------------ | |||
Bands : 100 | |||
K-points : 14 | |||
G-vectors [RL space]: 8029 | |||
Components [wavefunctions]: 1016 | |||
... | |||
[RD./SAVE//ns.wf]------------------------------------------- | |||
Fragmentation :yes | |||
... | |||
[RD./SAVE//ns.kb_pp_pwscf]---------------------------------- | |||
Fragmentation :yes | |||
- S/N 006626 -------------------------- v.04.01.02 r.00000 - | |||
In practice we suggest to move the <code>SAVE</code> into a new clean folder. | |||
In this tutorial however, we ask instead that you continue using a <code>SAVE</code> we prepared previously: | |||
$ cd ../../YAMBO | |||
$ ls | |||
SAVE | |||
== | == Links == | ||
* [[Advanced usage|Advanced usage of p2y]] | |||
* [[Tutorials|Back to tutorials menu]] | |||
Revision as of 13:43, 23 March 2017
In this tutorial you will learn how to generate the Yambo SAVE folder for bulk hBN starting from a PWscf calculation.
Prerequisites
You will need:
- PWSCF input files and pseudopotentials for hBN bulk (Download here)
pw.xexecutable, version 5.0 or laterp2yexecutable
System characteristics
Hexagonal boron nitride - hBN:
- HCP lattice, ABAB stacking
- Four atoms per cell, B and N (16 electrons)
- Lattice constants: a = 4.716 [a.u.], c/a = 2.582
- Plane wave cutoff 40 Ry (1500 RL vectors in wavefunctions)
- SCF run: shifted 6x6x6 grid with 8 bands
- Non-SCF run: unshifted 6x6x2 grid with 100 bands
DFT calculations
Unpack the tarfile. It uses the same file structure as other yambo tutorials:
$ tar -xcvf hBN-bulk.tar $ cd YAMBO_TUTORIALS/hBN/PWSCF $ ls Inputs Pseudos PostProcessing References hBN_scf.in hBN_nscf.in hBN_scf_plot_bands.in hBN_nscf_plot_bands.in
First run the SCF calculation to generate the ground-state charge density, occupations, Fermi level, and so on:
pw.x < hBN_scf.in > hBN_scf.out
The output reports 36 k-points. The valence band maximum is at 5.13eV.
Next run a non-SCF calculation to generate a set of Kohn-Sham eigenvalues and eigenvectors across a denser k-point mesh, for occupied and unoccupied states:
pw.x < hBN_nscf.in > hBN_nscf.out
Note the presence of the following flags in the input file:
wf_collect=.true. force_symmorphic=.true.
which are needed for generating the Yambo databases. Full explanations of these variables are given on the quantum-ESPRESSO input variables page.
After these two runs, you should have a hBN.save directory:
$ ls hBN.save data-file.xml charge-density.dat gvectors.dat B.pz-vbc.UPF N.pz-vbc.UPF K00001 K00002 .... K00035 K00036
Conversion to Yambo format
The PWscf bBN.save output is converted to the Yambo format using the p2y executable (pwscf to yambo), found in the yambo bin directory.
Enter hBN.save and launch p2y:
$ cd hBN.save $ p2y ... <---> DBs path set to . <---> Index file set to data-file.xml <---> Header/K-points/Energies... done ... <---> == DB1 (Gvecs and more) ... <---> ... Database done <---> == DB2 (wavefunctions) ... done == <---> == DB3 (PseudoPotential) ... done == <---> == P2Y completed ==
This output repeats some information about the system and generates a SAVE directory:
$ ls SAVE ns.db1 ns.wf ns.kb_pp_pwscf ns.wf_fragments_1_1 ... ns.kb_pp_pwscf_fragment_1 ...
These files, with an n prefix, indicate that they are in netCDF format, and thus not human readable. However, they are perfectly transferable across different architectures.
You are now ready to run Yambo. Check that the databases contain the information you expect:
$ yambo -D [RD./SAVE//ns.db1]------------------------------------------ Bands : 100 K-points : 14 G-vectors [RL space]: 8029 Components [wavefunctions]: 1016 ... [RD./SAVE//ns.wf]------------------------------------------- Fragmentation :yes ... [RD./SAVE//ns.kb_pp_pwscf]---------------------------------- Fragmentation :yes - S/N 006626 -------------------------- v.04.01.02 r.00000 -
In practice we suggest to move the SAVE into a new clean folder.
In this tutorial however, we ask instead that you continue using a SAVE we prepared previously:
$ cd ../../YAMBO $ ls SAVE