Quasi-particles of a 2D system: Difference between revisions
No edit summary |
No edit summary |
||
| Line 159: | Line 159: | ||
This will create a new input file and run it. The calculation databases and the human-readable files will be put in separate directories. Check the location of the report | This will create a new input file and run it. The calculation databases and the human-readable files will be put in separate directories. Check the location of the report <code>r-*</code> file and the log <code>l-*</code> files, and inspect them while the calculation runs. | ||
For simplicity you could just type | For simplicity you could just type | ||
tail -f run_MPI16_OMP1.out/LOG/l-*_CPU_1 | tail -f run_MPI16_OMP1.out/LOG/l-*_CPU_1 | ||
to monitor the progress in the master thread ( | to monitor the progress in the master thread (<code>Ctrl+C</code> to exit). | ||
As you can see, the run takes some time, even though we are using minimal parameters. | As you can see, the run takes some time, even though we are using minimal parameters. | ||
Meanwhile, we can run other jobs increasing the parallelisation. Let's employ 128 CPUs (i.e., half a CPU node on Vega). We'll use 128 MPI tasks, still with a single openMP thread. To this end modify the | Meanwhile, we can run other jobs increasing the parallelisation. Let's employ 128 CPUs (i.e., half a CPU node on Vega). We'll use 128 MPI tasks, still with a single openMP thread. To this end modify the <code>job_parallel.sh</code> script changing | ||
#!/bin/bash | |||
#SBATCH --nodes=1 | |||
#SBATCH --ntasks-per-node=128 | |||
#SBATCH --cpus-per-task=1 | |||
This time the code should be much faster. Once the run is over, try to run the simulation also on 32 and 64 MPI tasks. Each time, change <code>ntasks-per-node</code> to this number. Finally, you can try to produce a scaling plot. | |||
The timings are contained in the <code>r-*</code> report files. You can already have a look at them typing | |||
grep Time-Profile run_MPI*/r-* | |||
The python script [ | The python script [<code>parse_ytiming.py</code>](https://hackmd.io/@palful/HkU0xblHj#parse_ytiming.py) is useful for the post-processing of report files. You can already find it in the directory, together with the reports for the longer calculations with 1, 2, 4 and 8 MPI tasks which have been provided. | ||
If you didn't do so already, load the python module | If you didn't do so already, load the python module ???? | ||
``` | ``` | ||
module purge | module purge | ||
| Line 196: | Line 193: | ||
python parse_ytiming.py run_MPI | python parse_ytiming.py run_MPI | ||
``` | ``` | ||
to look for a report file in each | to look for a report file in each <code>run_MPI*.out</code> folder. Make sure you have only one report file per folder. | ||
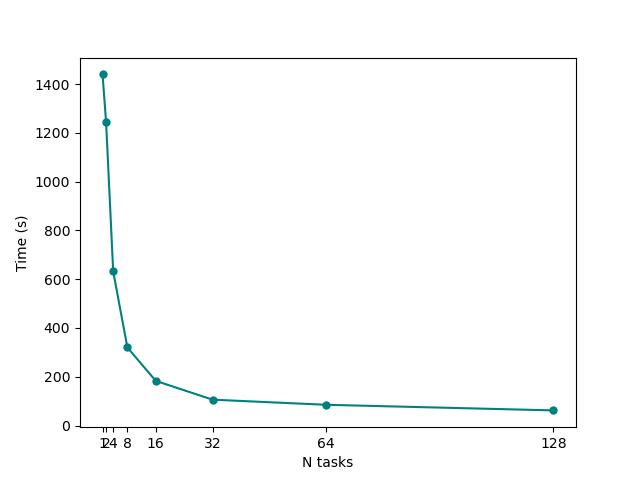
You can also play with the script to make it print detailed timing information, however you should already see that it produced a png plot showing times-to-completion on y axis against number of MPI tasks on the x axis. | You can also play with the script to make it print detailed timing information, however you should already see that it produced a png plot showing times-to-completion on y axis against number of MPI tasks on the x axis. | ||
[[File:Time parall.png|thumb]] | |||
What can we learn from this plot? In particular, try to answer the following questions: | What can we learn from this plot? In particular, try to answer the following questions: | ||
Revision as of 15:36, 7 May 2023
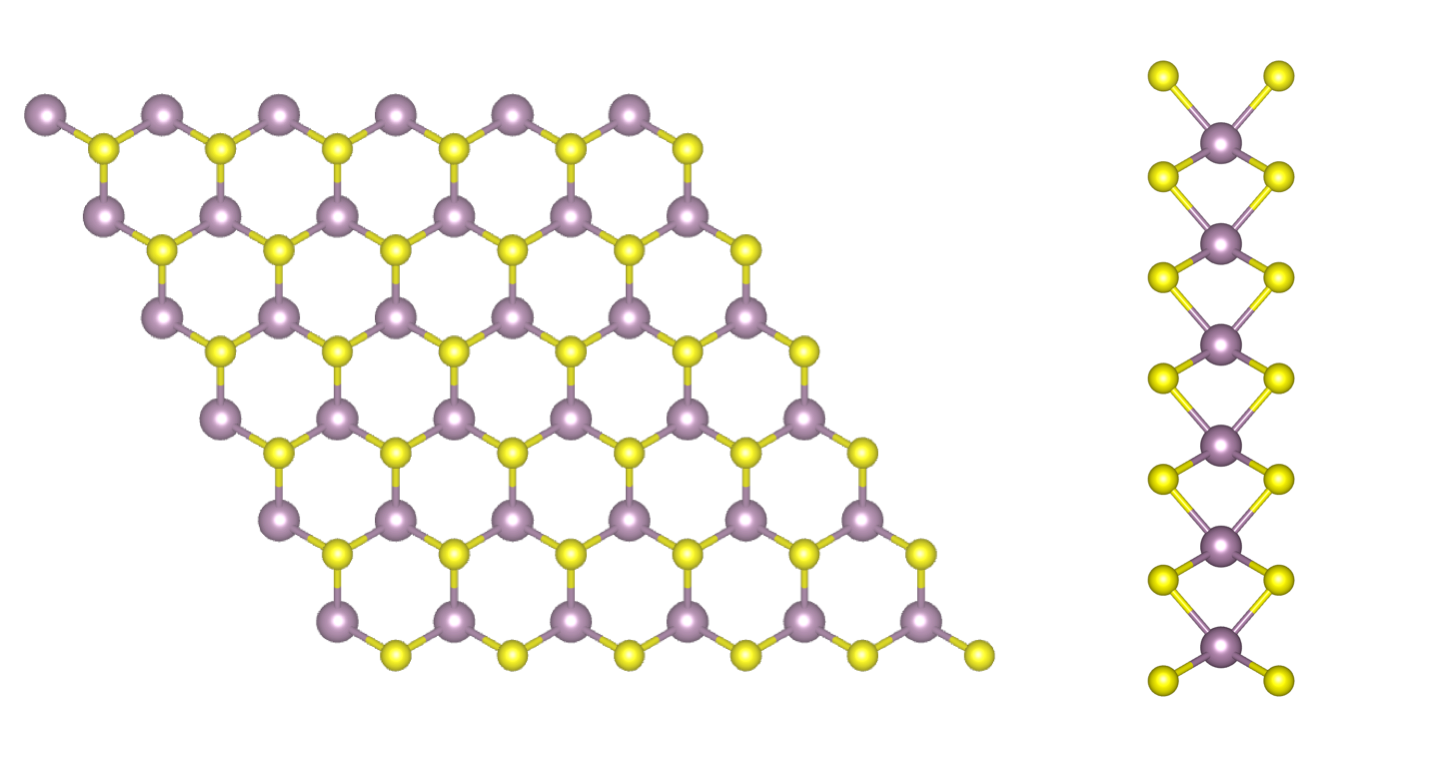
In this tutorial you will compute the quasiparticle corrections to the band structure of a free-standing single layer of MoS2. Aim of the tutorial is to learn how to efficiently run a GW simulation in a 2D material based on:
- Acceleration techniques of GW in 2D systems
- Parallelization techniques
In the end, you will obtain a quasiparticle band structure based on the simulations, the first step towards the reproduction of an ARPES spectrum. Beware: we won’t use fully converged parameters, so the final result should not be considered very accurate.
Step 1: Initialization
The first step would be as usual to perform a non-self-consistent DFT calculation of MoS2 (using for example Quantum ESPRESSO). Here, you can directly download the QE files from [1].
Once completed the extraction, move inside the 00_QE-DFT folder and then convert QE data into a convenient format for Yambo. Just as a reminder, to convert, run the p2y executable to generate the uninitialised SAVE.
cd 00_QE-DFT/mos2.save p2y
Now, we need to run the initialization step. Every Yambo run **must** start with this step. Just type
yambo
==Step 2: Use the RIM-W accelerator__
<details>
However you can try to get a reasonable correction via the RIM-W approach. ``` cp i02-GW_80_Gbands i04-GW_80_Gbands_rimw vim i04-GW_80_Gbands_rimw ``` Then, you just need to add the following two variables to the input file ```bash= RIM_W RandGvecW= 15 RL ``` and prepare a submission script ``` cp run02_converge_Gbnds.noBG.sh run04_converge_rimw.sh vim run04_converge_rimw.sh ``` and edit it, by removing the loop and changin the input file and jobname ```bash=
- !/bin/bash
- SBATCH --nodes=1
- SBATCH --ntasks-per-node=16
- SBATCH --cpus-per-task=1
- SBATCH --partition=cpu
- SBATCH --time=0:30:00
- SBATCH --account=d2021-135-users
- SBATCH --mem-per-cpu=2000MB
- SBATCH --job-name=mos2
- SBATCH --reservation=maxcpu
- load yambo and dependencies
module purge module use /ceph/hpc/data/d2021-135-users/modules module load YAMBO/5.1.1-FOSS-2022a export OMP_NUM_THREADS=${SLURM_CPUS_PER_TASK}
summaryfile=summary_04_rimw.txt
echo 'E_vale [eV] E_cond [eV] GAP [eV]' > $summaryfile
G_BANDS=80
label=Gbands_${G_BANDS}_rimw jdir=job_${label} cdir=out_${label} filein=i04-GW_${G_BANDS}_Gbands_rimw
- run yambo
srun --mpi=pmix -n ${SLURM_NTASKS} yambo -F $filein -J $jdir -C $cdir
E_GW_v=`grep -v '#' ${cdir}/o-${jdir}.qp|head -n 1| awk '{print $3+$4}'` E_GW_c=`grep -v '#' ${cdir}/o-${jdir}.qp|tail -n 1| awk '{print $3+$4}'`
GAP_GW=`echo $E_GW_c - $E_GW_v |bc`
echo ${E_GW_v} ' ' ${E_GW_c} ' ' ${GAP_GW} >> $summaryfile ``` and then run ``` sbatch run04_converge_rimw.sh ``` How much do you get for the band gap ? </details>
Step 3: GW parallel strategies
MPI parallelization
For this section, let us enter the 03_GW_parallel directory. If you were in the 02_GW_convegence folder just type
cd ../03_GW_parallel
and inspect the input gw.in. You will see that we set low values for most of the convergence parameters except bands:
vim gw.in
FFTGvecs= 20 Ry # [FFT] Plane-waves
EXXRLvcs= 2 Ry # [XX] Exchange RL components
VXCRLvcs= 2 Ry # [XC] XCpotential RL components
% BndsRnXp
1 | 300 | # [Xp] Polarization function bands
%
NGsBlkXp= 1 Ry # [Xp] Response block size
% GbndRnge
1 | 300 | # [GW] G[W] bands range
%
%QPkrange # [GW] QP generalized Kpoint/Band indices
1| 19| 23| 30|
%
Note that we also added the quantity FFTGvecs to reduce the size of the Fourier transforms (the default corresponds to Quantum ESPRESSO ecutwfc, being 60 Ry in this case).
In addition, we have deleted all the parallel parameters since we will be setting them via the submission script.
Actually we are now dealing with a heavier system than before: as you can see from the QPkrange values, we have switched to a 12x12x1 k-point grid - having 19 points in the irreducible Brillouin zone - and turned spin-orbit coupling on in the DFT calculation - now the top valence band is number 26 instead of 13 because the bands are spin-polarized.
The inspected file should look like [this one ????](https://hackmd.io/@palful/HkU0xblHj#GW-input-file-for-parallel-section).
For this part of the tutorial, we will be using the slurm submission script [`job_parallel.sh`](https://hackmd.io/@palful/HkU0xblHj#Slurm-submission-script-on-VEGA), which is available in the calculation directory.
If you inspect it, you will see that the script adds additional variables to the yambo input file.
These variables control the parallel execution of the code:
DIP_CPU= "1 $ncpu 1" # [PARALLEL] CPUs for each role DIP_ROLEs= "k c v" # [PARALLEL] CPUs roles (k,c,v) DIP_Threads= 0 # [OPENMP/X] Number of threads for dipoles X_and_IO_CPU= "1 1 1 $ncpu 1" # [PARALLEL] CPUs for each role X_and_IO_ROLEs= "q g k c v" # [PARALLEL] CPUs roles (q,g,k,c,v) X_and_IO_nCPU_LinAlg_INV= 1 # [PARALLEL] CPUs for Linear Algebra X_Threads= 0 # [OPENMP/X] Number of threads for response functions SE_CPU= " 1 $ncpu 1" # [PARALLEL] CPUs for each role SE_ROLEs= "q qp b" # [PARALLEL] CPUs roles (q,qp,b) SE_Threads= 0 # [OPENMP/GW] Number of threads for self-energy
The keyword DIP refers to the calculations of the screening matrix elements (also called "dipoles") needed for the screening function, X is the screening function (it stands for [math]\displaystyle{ \chi }[/math] since it is a response function), SE the self-energy.
These three sections of the code can be parallelised independently.
- info
- In this subsection we are mainly concerned with the **[PARALLEL]** variables which refer to MPI _tasks_. - We will see later an example of **[OPENMP]** parallelisation (i.e., adding _threads_).
We start by calculating the QP corrections using 16 MPI tasks and a single openMP thread. Therefore, edit the submission script as:
#!/bin/bash #SBATCH --nodes=1 #SBATCH --ntasks-per-node=16 #SBATCH --cpus-per-task=1
and submit the job
sbatch job_parallel.sh
This will create a new input file and run it. The calculation databases and the human-readable files will be put in separate directories. Check the location of the report r-* file and the log l-* files, and inspect them while the calculation runs.
For simplicity you could just type
tail -f run_MPI16_OMP1.out/LOG/l-*_CPU_1
to monitor the progress in the master thread (Ctrl+C to exit).
As you can see, the run takes some time, even though we are using minimal parameters.
Meanwhile, we can run other jobs increasing the parallelisation. Let's employ 128 CPUs (i.e., half a CPU node on Vega). We'll use 128 MPI tasks, still with a single openMP thread. To this end modify the job_parallel.sh script changing
#!/bin/bash #SBATCH --nodes=1 #SBATCH --ntasks-per-node=128 #SBATCH --cpus-per-task=1
This time the code should be much faster. Once the run is over, try to run the simulation also on 32 and 64 MPI tasks. Each time, change ntasks-per-node to this number. Finally, you can try to produce a scaling plot.
The timings are contained in the r-* report files. You can already have a look at them typing
grep Time-Profile run_MPI*/r-*
The python script [parse_ytiming.py](https://hackmd.io/@palful/HkU0xblHj#parse_ytiming.py) is useful for the post-processing of report files. You can already find it in the directory, together with the reports for the longer calculations with 1, 2, 4 and 8 MPI tasks which have been provided.
If you didn't do so already, load the python module ???? ``` module purge module load matplotlib/3.2.1-foss-2020a-Python-3.8.2 ```
Then, after your jobs have finished, run the script as
```
python parse_ytiming.py run_MPI
```
to look for a report file in each run_MPI*.out folder. Make sure you have only one report file per folder.
You can also play with the script to make it print detailed timing information, however you should already see that it produced a png plot showing times-to-completion on y axis against number of MPI tasks on the x axis.
What can we learn from this plot? In particular, try to answer the following questions:
- Up to which number of MPI tasks our system efficiently scales? At which point adding more threads starts to become a waste of resources?
- How could we change our parallelisation strategy to improve the scaling even more?
- info
Keep in mind that the MPI scaling we are seeing here is not the true yambo scaling, but depends on the small size of our tutorial system. In a realistic calculation for a large-sized system, __yambo has been shown to scale well up to tens of thousands of MPI tasks__!
- Hybrid parallelization strategy
It turns out that for this system it's not a good idea to use more than 32 MPI tasks. But we still have 128 CPUs available for this run. How to optimize the calaculation even more? We can try using OpenMP threads.
This is turned on by modifying `cpus-per-task` in the submission file. The product of the number of OpenMP threads and MPI tasks is equal to the total number of CPUs. Therefore, we can distribute 128 cpus with 32 MPI tasks (efficient MPI scaling, see above) and then use 4 OpenMP threads:
```bash=
- !/bin/bash
- SBATCH --nodes=1
- SBATCH --ntasks-per-node=32
- SBATCH --cpus-per-task=4
```
Actually, we don't need to change the related openMP variables for the yambo input, since the value `0` means "use the value of `OMP_NUM_THREADS`" and we have now set this environment variable to `4` via our script. Otherwise, any positive number can directly specify the number of threads to be used in each section of the code.
```bash= DIP_Threads= 0 # [OPENMP/X] Number of threads for dipoles ... X_Threads= 0 # [OPENMP/X] Number of threads for response functions ... SE_Threads= 0 # [OPENMP/GW] Number of threads for self-energy ```
You can try to run this calculation and check if it is faster than the pure 128 MPI one from before. You should find a massive gain: in our case, the pure `MPI128_OMP1` took 01m-02s, while the hybrid `MPI32_OMP4` took just 37s, a 40% speedup.
You can also try any other thread combinations including the pure openMP scaling, and compare the timings.
- info
- In real-life calculations running on $n_{cores} > 100$, as we have seen, it may be a good idea to adopt a hybrid approach. - The most efficient scaling can depend both on your system and on the HPC facility you're running on. For a full CPU node on Vega (256 cores), using a large-scale system, we have found that 16 tasks times 16 threads gives the best performance. - OpenMP can help lower memory requirements within a node. You can try to increase the OpenMP share of threads if getting Out Of Memory errors.
As mentioned above, we can conclude that the "best" parallelisation strategy is in general both system-dependent and cluster-dependent. If working on massive systems, it may be well worth your while to do preliminary parallel tests in order to work out the most efficient way to run these jobs.
- [OPTIONAL]: Comparing different parallelisation schemes
<details>
Up to now we always parallelised over a single parameter, i.e. `c` or `qp`. However, Yambo allows for tuning the parallelisation scheme over several parameters broadly corresponding to "loops" (i.e., summations or discrete integrations) in the code.
To this end you can open again the `run_parallel.sh` script and modify the section where the yambo input variables are set.
For example, `X_CPU` sets how the MPI Tasks are distributed in the calculation of the response function. The possibilities are shown in the `X_ROLEs`. The same holds for `SE_CPU` and `SE_ROLEs` which control how MPI Tasks are distributed in the calculation of the self-energy.
```bash= X_CPU= "1 1 1 $ncpu 1" # [PARALLEL] CPUs for each role X_ROLEs= "q g k c v" # [PARALLEL] CPUs roles (q,g,k,c,v) X_nCPU_LinAlg_INV= $ncpu # [PARALLEL] CPUs for Linear Algebra X_Threads= 0 # [OPENMP/X] Number of threads for response functions DIP_Threads= 0 # [OPENMP/X] Number of threads for dipoles SE_CPU= "1 $ncpu 1" # [PARALLEL] CPUs for each role SE_ROLEs= "q qp b" # [PARALLEL] CPUs roles (q,qp,b) SE_Threads= 0 # [OPENMP/GW] Number of threads for self-energy ```
You can try different parallelization schemes and check the performances of Yambo. In doing so, you should also change the jobname `label=MPI${ncpu}_OMP${nthreads}` in the `run_parallel.sh` script to avoid confusion with previous calculations.
You may then check how speed, memory and load balance between the CPUs are affected. You could modify the script [`parse_ytiming.py`](https://hackmd.io/@palful/HkU0xblHj#parse_ytiming.py) to parse the new data, read and distinguish between more file names, new parallelisation options, etc.
- info
- The product of the numbers entering each variable (i.e. `X_CPU` and `SE_CPU`) times the number of threads should always match the total number of cores (unless you want to overload the cores taking advantage of multi-threads). - Using the `X_Threads` and `SE_Threads` variables you can think about setting different hybrid schemes between the screening and the self-energy runlevels. - Memory scales better if you parallelize on bands (`c v b`). - Parallelization on $k$-points performs similarly to parallelization on bands, but requires more memory. - Parallelization on $q$-points requires much less communication between the MPI tasks. It may be useful if you run on more than one node and the inter-node connection is slow.